Goals
Myself and a group of researchers were interested developing personalized nutrition plans by better understanding how diet could uniquely affect each individuals diet. We were tackling this problem by applying machine learning methods to dietary information and continuous blood sugar data. While there were many automated approaches for labeling blood sugar readings as either "good" or "bad", we wanted clinical-grade labels for validating these machine learning models.
Users
Clinician and data scientist. A clinician, experienced with assessing continuous blood sugar data, was the one to use the web application and annotate the data. The data scientist (in this case, myself) was the one to utilize the annotated data to build the machine learning models.
Problem
The initial method of annotation required too much context switching and was too time consuming to obtain sufficient amounts of labeled data for validation. The continuous blood sugar data was stored in CSV files, which required the clinician to build a graph for each record in excel and mark their notes in another document. A single annotation took 2-5 minutes and the project required 50-100 annotations for minumal validation when the clinician needed to be spending their time seeing patients. With this approach, building a clinician-labeled dataset was not possible.
Solution
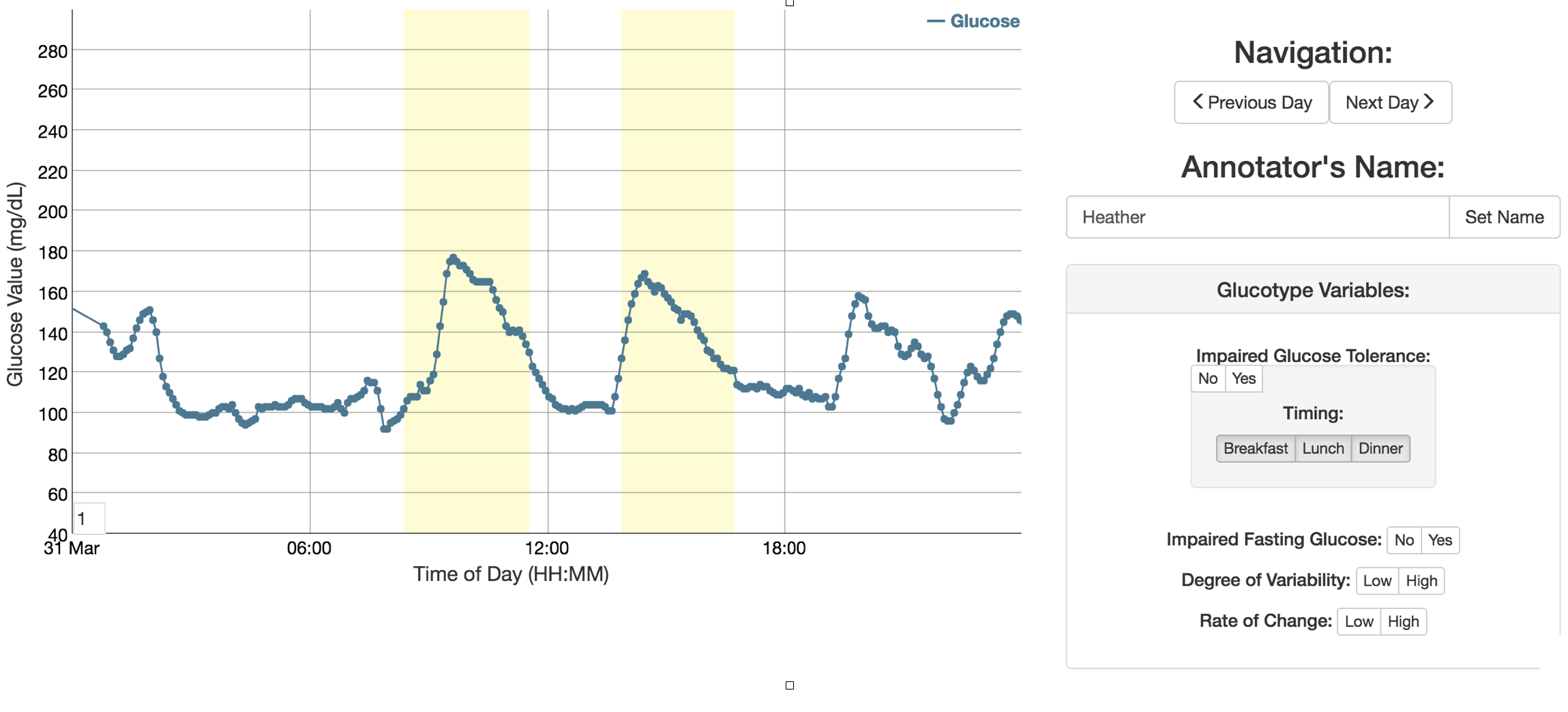
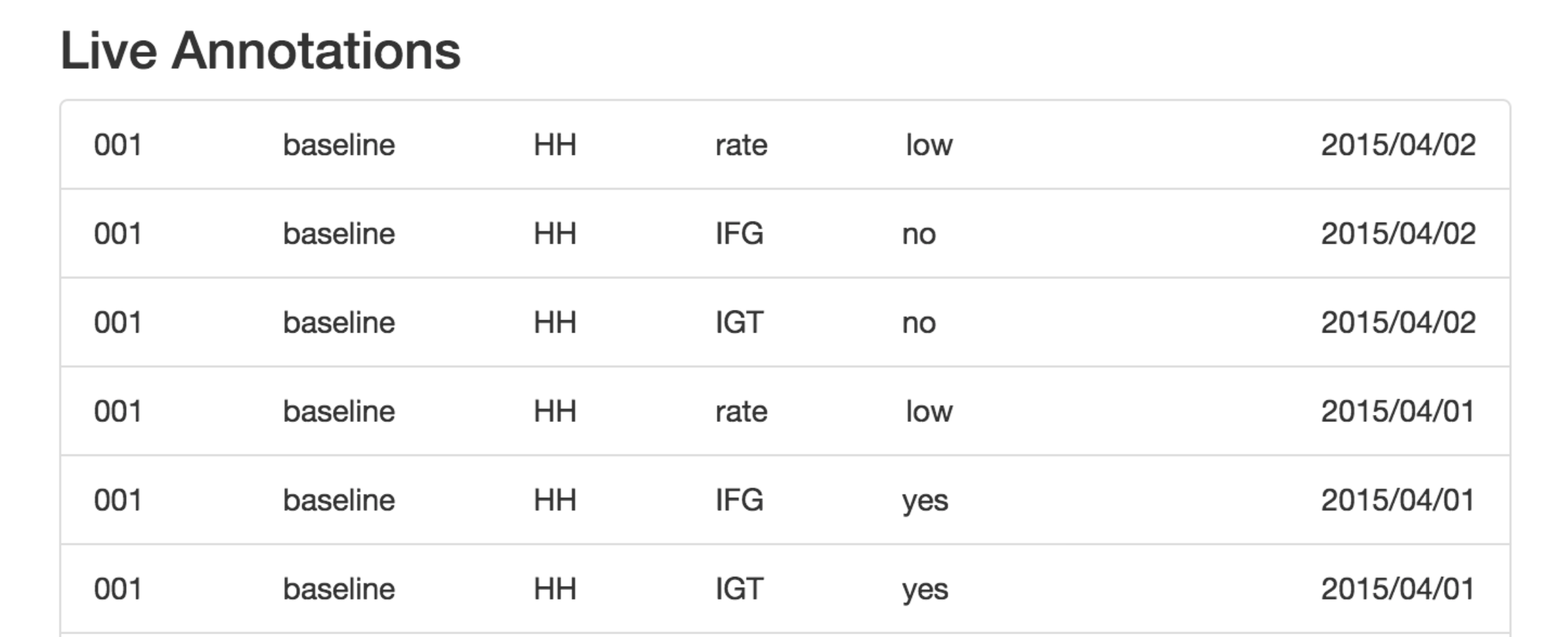
I built an application that allowed the clinician to browse the continuous blood sugar data by patient or view randomly for annotation. The continuous blood sugar data was displayed on a per day basis and enabled the clinician to submit annotations for storaged in an SQL database. The clinician could highlight "bad" blood sugar events that would be stored with a start and end times. In addition, the clinician could add labeled that were predetermined to be of interest for the machine learning model. These annotations could then be queried and easily formatted for machine learning and validation purposes.
In order to develop the interface for the web application, the clinican and I discussed how to describe a "poor" blood sugar response and what information she needed to make her assessments. We then translated these to keywords and levels that would be most relevant for validation of the machine learning models. As the data scientist, I determined the final format in which to store these annotations in the database. I then worked with a web developer to build an MVP that would meet the minimal requirements for the annotation. Since I was the data scientist for the project, the intricacies of the data and endgoals were well known. After several iterations to focus the clinicians efforts on only those vital annotations for the project, we sent her the website to begin annotation. Within a few days, we had sufficient labled data and were able to perform the validation of the machine learning models.
- Skills : Web development, Dygraph.js, SQL database
- Category : Data Visualization, Softweare Engineering, Design
- Date : 2015
